Graybill
Conference 2003
PROGRAM
June 18-20, 2003
Colorado
State University Campus
and
University Park Holiday Inn
Fort Collins, CO
Wednesday, June 18
8:00 AM to 8:30 AM Room 206, Weber Building, CSU campus
Coffee, juice, and donuts
8:30 AM – 4:30 PM Room 206, Weber Building, CSU campus
Tutorial on “Analysis of DNA Microarray Data” by Dr. Steen Knudsen, Associate Professor, Deputy Director, Center for Biological Sequence Analysis, Technical University of Denmark
Lunch break from NOON to
1:30 PM
7:00
PM Registration Packet Pickup
University Park Holiday Inn – Oklahoma State Room
7:00
PM to 9:00 PM Conference
Opening mixer
University Park Holiday Inn – Oklahoma State Room
Thursday,
June 19
7:30 AM Registration Packet Pickup
University Park Holiday Inn – Oklahoma State Room
IDAHO
STATE – MICHIGAN STATE ROOM
8:00 AM Welcome address and opening remarks
8:15 AM Session I Session Chair: Phillip Chapman
8:15 AM to 8:45 AM
David Allison
Statistical Genetics, University of Alabama-Birmingham
Applying High-Dimensional Approaches to Microarray Research
8:45 AM to 9:15 AM
Rebecka Jornsten
Department of Statistics, Rutgers University
Clustering, Classification and Validation via the L1 Data Depth
9:15 AM to 9:45 AM
Grace Shieh
Institute of Statistical Science, Academia Sinica, Taipei, Taiwan
Reconstructing Yeast Gene Networks from Microarray
Data
9:45 AM
to 10:15 AM
BREAK – ATRIUM
10:15 AM Session II Session Chair: Jim zumBrunnen
10:15 AM to 10:45 AM
Lue Ping Zhao
Division of Public Health Sciences, Fred Hutchinson Cancer Research Center
Building a Predictive Model Using Array Technologies with
an Application to Breast Cancer
10:45 AM to 11:15 AM
Lawrence Hunter
Center for Computational Pharmacology, University of Colorado Health Sciences Center
Knowledge-based Interpretation of Gene Expression Array Studies
Keynote Address
11:15 AM to 12:15 PM
Russell
D. Wolfinger
SAS
Institute, Inc.
Extending Graybill's Legacy
to Microarray Data and Beyond
12:15 PM to 1:30 PM
LUNCH at the Holiday Inn ATRIUM
1:30 PM
Session
III
Session Chair: Thomas Lee
1:30 PM to 2:00 PM
Jane Chang
Dept. of Applied Statistics and Operations Research, Bowling Green State University
Design and Analysis of 2-Channel Microarray Experiments
2:00 PM to 2:30 PM
Chen-Tuo Liao
Division of Biometry, Institute of Agronomy, National Taiwan University
Statistical Designs for cDNA Microarray Experiments
2:30 PM to 3:00 PM
Susan Hilsenbeck
Baylor College of Medicine, Texas Medical Center
Pilot Studies of Components of Technical Variability and Practical Recommendations for Study Design
3:00 PM
to 3:30 PM
BREAK – ATRIUM
3:30 PM
Session IV
Session
Chair: Thomas Boardman
STUDENT PRESENTATIONS
3:30 PM to 3:50 PM
Deepak Mav
Department of Mathematics and Statistics, Old Dominion University
Bivariate
Models for Identifying Differentially Expressed Genes in Microarray Experiments
3:50 PM to 4:10 PM
Annette Molinaro
Division of Biostatistics, University of California at Berkeley
Prediction of Survival with Regression Trees and Cross-Validation
4:10 PM to 4:30 PM
Gail R. Willsky1,
Lai-Har Chi1 , Yulan
Liang2 and
Debbie C Crans3
University at Buffalo (SUNY) Dept. of Biochemistry1 and Division of Statistics, Dept. of Social and Preventative Medicine2 and Colorado State University, Dept. of Chemistry3
Identification of Gene Expression Changes in Skeletal Muscle from Diabetic and Normal Rats Treated by Oral Administration of Vanadyl Sulfate
6:30 PM Pre-banquet Social and Cash Bar - Oklahoma State Room
7:00 PM Banquet – Oklahoma State Room
Guest
Speaker: Robert V.
Hogg Professor
Emeritus, University of Iowa
Friday, June 20
8:00 AM
SESSION V
Session
Chair: Jan Hannig
8:00 AM to 8:30 AM
Wei Pan
Biostatistics, University of Minnesota
Statistical Significance Analysis of Longitudinal Gene Expression Data
8:30 AM to 9:00 AM
Dan Nettleton
Department of Statistics, Iowa State University
Identifying Differentially Expressed Genes in Unreplicated Multiple-Treatment Time-Course Microarray Experiments
9:00 AM to 9:30 AM
Peter Munson
Mathematical and Statistical Computing Lab, National Institutes of Health
Significance of Gene Expression Changes Using the "Consistency" Test
9:30 AM to 10:00 AM
Laura Lazzeroni
Biostatistics and Genetics, Stanford University
Results from the Distribution of the Number of False Positives: Examples from Microarrays
10:00
AM to 10:30 AM
BREAK – ATRIUM
10:30
AM Session VI
Session Chair: Jay Breidt
10:30 AM to 11:00 AM
David M. Rocke
Department of Applied Science and Division of Biostatistics, University of California-Davis
Measurement
Errors and Data Transformation for Gene Expression Data, Proteomics Data, and
Metabolomics Data
11:00 AM to 11:30 AM
Jenny Bryan
Biotechnology Laboratory & Department of Statistics, University of British Columbia
Resampling to Assess Significance of Gene Clusters
11:30 AM to 12:00 Noon
Steen Knudsen
Center for Biological Sequence Analysis, Technical University of Denmark
Bioinformatics Analysis of DNA Microarray Data
12:00 NOON to 1:15 PM LUNCH at the Holiday Inn ATRIUM
1:15 PM
Session VII
Session Chair: Hari Iyer
1:15 PM to 1:45 PM
Philip Dixon
Department of Statistics, Iowa State University
Modeling Nucleotide Sequence Variation in a Viral
Quasi Species
1:45 PM to 2:15 PM
Kent Eskridge
Biometry, University of Nebraska
Field Design and Analysis in the Search for Quantitative Trait Loci in
Plants
2:15 PM to 2:45 PM
Deborah Glueck
Department of Preventive Medicine and Biometrics, Univ. of Colorado Health Sciences Center
Exact
Power Under Independence for the False Discovery Rate in Gene Expression Array
Experiments
2:45 PM to 3:15 PM
David L. Gold
M. D. Anderson Cancer Center, The University of Texas
Graphically Displaying Partitions of Variation in Microarray Data
3:15 PM CLOSING REMARKS
3:20 PM
BREAK -- FAREWELL MIXER -- ATRIUM
|
Map of Fort Collins Area |
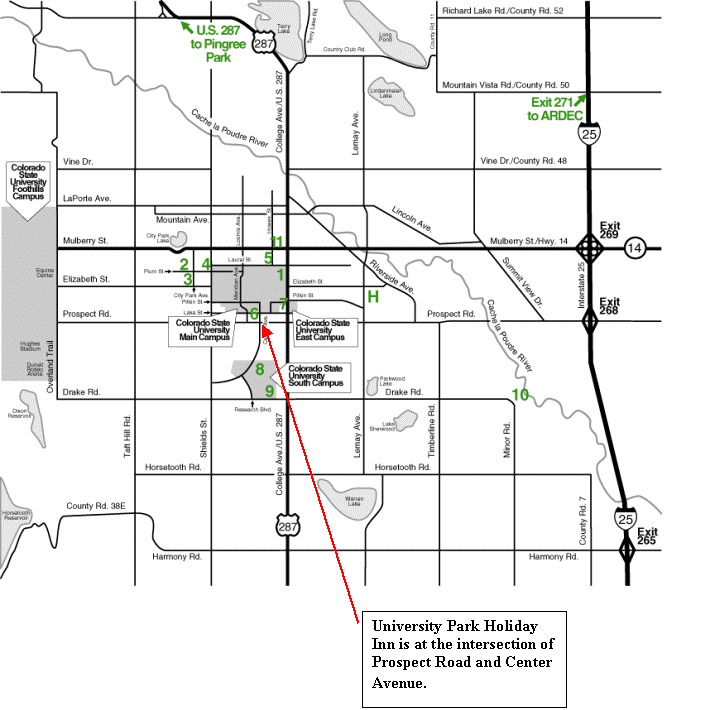
|
|
Map of Fort Collins Area |
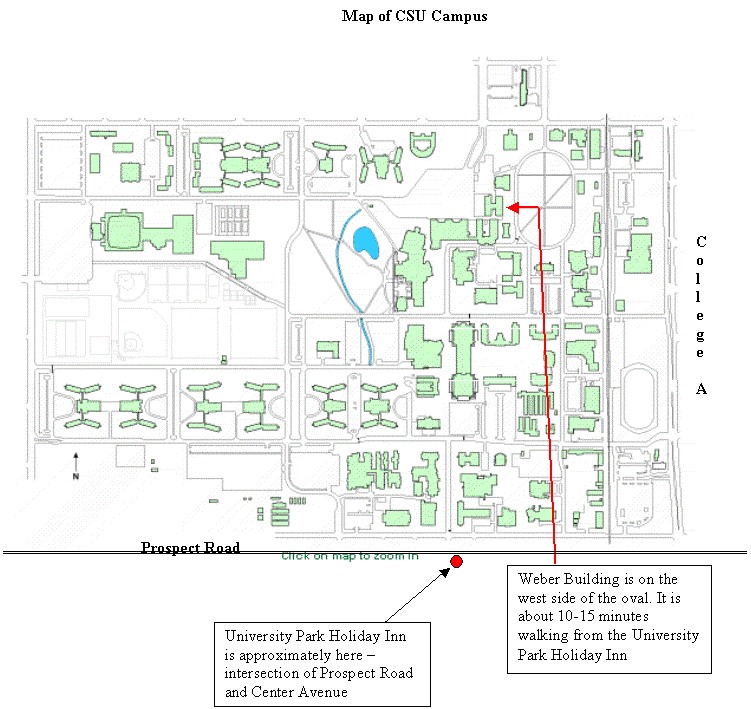
|